Epigenetics and Gastrointestinal Microbiomes Influence on the Gut-Brain Axis and its Effect on Disease Causation and Prevention.
By Dustin Moffitt, ND, 2015
Copyrighted by: Prospectively Healthy ©
Abstract
As we all know as laboratory diagnostic interpretations improve and we not only know more what markers to look for when testing, but also correlate what the values may determine, leading to new trends in healthcare or science in general. Many practitioners and researchers are becoming aware of the influence of our gastrointestinal (GI) microbiome and its influence upon the brains cognitive function, but few have realized that the brain also greatly influences the microbiome itself. Many factors may affect this intercommunication pathway ranging from stress, injury, emotion, dietary habits and even our environmental surroundings. These can alter the tolerance of the GI tract including the epithelial barrier, GI motility, nutrient and byproduct absorption and secretion, as well as the enteric nervous system. All the data presented here is a compilation of many studies and interpreted in a way to pave the pathway to new research to gain a better understanding of the relationship between lifestyle choices and behavior and their influence on the GI microbiome, and vice versa. This will be an ongoing research protocol for myself, so please stick with me as it grows.
Keywords: microbiota, gut-brain axis, prebiotics, probiotics, epigenetics
What is the Gut Brain Axis?
The gut-brain axis (GBA) is a bidirectional signaling mechanism that exists between the gastrointestinal (GI) tract and the brain, integrating the functions crucial to maintaining homeostasis via hormonal, immunological, and neurological communications.1 As the GBA becomes disrupted, we see the GI bacterial balance start to stray from its normal colonies. Hence the GBA has been highly attributed to gut dysbiosis and inflammatory bowel disease2 (IBD) following traumatic brain injuries (TBI).
Microbiome Influence on GI-tract
Although it is not entirely clear as to what exact mechanism is in place for the influence that the microbiome may play in the functionality of the brain, many current studies are occurring to continue gathering information. We do know that there are 1014 microorganisms living in the human body with a large number of them residing in the GI tract, with this the microbiome consist of 100 times more genes than that of the human genome, therefore the microflora has been referred to as the forgotten organ 3. Although the bacterial species vary distinctively between one person to the next
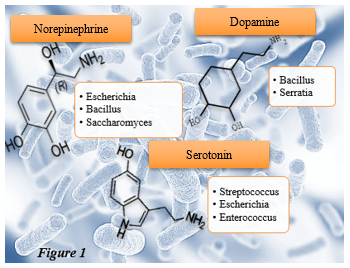
The primary phyla that the biome consists of are Bacteroidetes, Firmicutes, Actinobacteria, and Proteobacteria. Key neuroendocrine transmitters such as norepinephrine (NE), Serotonin (5-HT), and Dopamine (DA) have been linked to certain microorganisms.4 (Fig. 1) We also know that higher levels of Actinobacteria and Firmicutes, including lactobacilli, in the GI are indicators of a good marker for health. Generically speaking, those with autoimmune disease and obesity5 tend to have a decreased amount of these phyla.
GBA interactions
Generally speaking, we are more proactive in dealing with stress when our gut microflora is more balanced between commensals and opportunists. In a balanced microflora we see the release of metabolites that act as endocrines leading to neurotransmitter response mechanisms. This is done so through M cells presenting cells to immune system leading to increased dendritic cells response to invaders. An example being Proteobacteria gram (–) lipopolysaccharide that lead to localized inflammatory responses and in some cases, systemically.
Microbiota and the Immune System
The immune system shapes the microbiota by affecting the mucus layer, via secretion of antibacterial proteins and IgA. While the microbiota impacts the immune systems development via the formation of organized lymphoid tissue and regulation of innate lymphoid cells. CD4, Treg, Th17 cells. (Fig. 3)
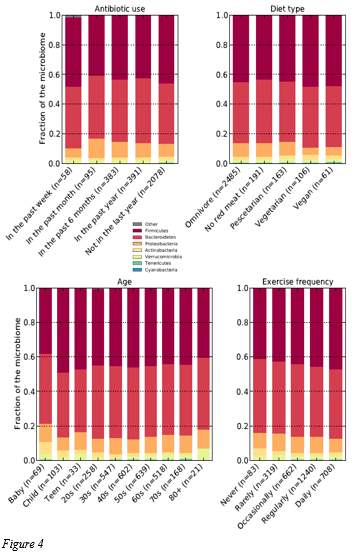
Epigenetic key influencing factor
Figure 4
Microbiome and GBA in Relation to Health and Disease
There are many research designs presented to the IRB each year involving the microbiota metagenomics.6 Currently there are HMP demonstration projects that involve diseases related to the microbiome that include; Crohn’s disease, acne, obesity, febrile illness, vaginal disorders, nutrigenomics, ulcerative colitis, esophageal adenocarcinoma, psoriasis, bacterial vaginosis, necrotizing enterocolitis, and intestinal inflammation.
Based on animal studies done, the microbiome in fact does influence the ability of the brain to function and even goes as far to modify our social behaviors.
Plant sources such as vegetables and fruits used for human consumption have been shown to lead to an increased number of beneficial microbiota and it is believed to be due to the plants containing endophytes. We also know that exercise, sleep, stress and other foods play a substantial role in this balance as well.
Gut Microbiome Testing
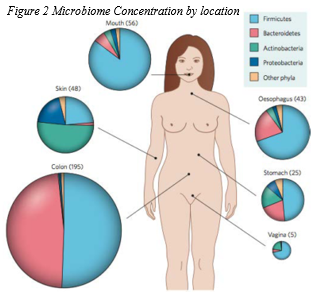
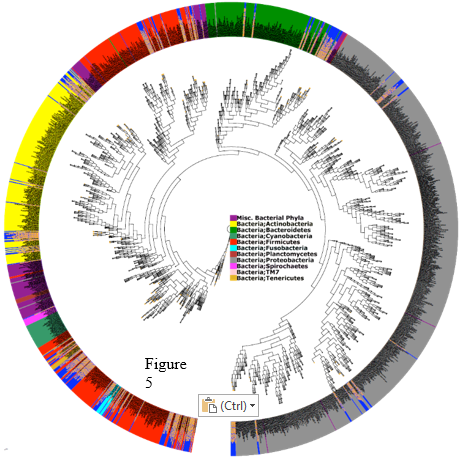
The microbiome has been tested for many years, however it was not until 2007 when that National Institutes of Health (NIH) launched the Human Microbiome Project (HMP). The first phase of HMP lasted from 2007 until 2012 and was responsible for developing metagenomics datasets and establishing tools for characterizing the microbiome in healthy adults and cohorts of specific microbiome-associated diseases. 7 As with the exponential number of variant organisms inhabiting GI in any given persons this leaves a large portion of the genome that has yet to still be isolated and sequenced, attributed to the inability to equally reproduce the growing conditions in a lab. Metagenomics, which is the study of microbial communities from environmental samples. 8 The HMP is using 16s rRNA and the metagenomic sequencing to map out the human microbiome in a total of 15-18 locations. Here is an example of some of the data being compiled, and a chart of the phyla collected (Fig 5.)9
As this compilation is primarily about that involving the GBA, we will only be focusing on the microbiome testing involving the GI tract. Currently there are two mainstream testing agencies that the public as well as practitioners have access to, being µBiome and American Gut. µBiome offers testing for a range of 1-5 sites and priced $89-$399. American Gut is offering testing from one to all areas and ranging from prices of $75-$25,000. These tests are all done at the subjects own will and financial obligation, as these are still elective laboratory interpretations. All of the data collected from these projects are being databased and mapped1> to divide into phyla and comparatively charted with the respective known diseases.
Laboratory diagnostic protocols must be followed to be represented in the HMP collection of data. The protocols in an abbreviated manor are testing the 454 16S to verify against each sample and the local and community databases, and against human contamination. The full protocol may be found here. Microbiome testing is still in the “experimental” phase, not because the testing is not adequate, but more so that it is not known fully how to interpret and clinically apply all the data collected. The more data that is added in the HMP database the greater the relevance of the collected database as we can start noticing trends of phyla or even particular bacterial organism in relation to health and disease.
References
- Collins SM, Bercik P. The Relationship Between Intestinal Microbiota and the Central Nervous System in Normal Gastrointestinal Function and Disease. Gastroenterology. 2009;136(6):2003-2014. doi:10.1053/j.gastro.2009.01.075.
- Tsaousides T, Gordon W a. Cognitive rehabilitation following traumatic brain injury: assessment to treatment. Mt Sinai J Med. 2009;76(2):173-181. doi:10.1002/MSJ.
- O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7(7):688-693. doi:10.1038/sj.embor.7400731.
- Lyte M. Microbial Endocrinology in the Microbiome-Gut-Brain Axis: How Bacterial Production and Utilization of Neurochemicals Influence Behavior. PLoS Pathog. 2013;9(11):e1003726. doi:10.1371/journal.ppat.1003726.
- Ley RE. Obesity and the human microbiome. Curr Opin Gastroenterol. 2010;26(1):5-11. doi:10.1097/MOG.0b013e328333d751.
- Human Gut Microbiome in Crohn’s Disease (ID 46879) – BioProject – NCBI. http://www.ncbi.nlm.nih.gov/bioproject/46321. Accessed April 22, 2015.
- National Institutes of Health. Human Microbiome Project. 2013. http://commonfund.nih.gov/hmp/. Accessed April 22, 2015.
- Lane DJ, Pace B, Olsen GJ, Stahl D a, Sogin ML, Pace NR. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A. 1985;82(20):6955-6959. doi:10.1073/pnas.82.20.6955.
- Li J, Jia H, Cai X, et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32(8):834-841. doi:10.1038/nbt.2942.
- Nelson KE, Weinstock GM, Highlander SK, et al. A catalog of reference genomes from the human microbiome. Science. 2010;328(5981):994-999. doi:10.1126/science.1183605.
Resources
- Ventura M, Canchaya C, Tauch A, et al. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev. 2007;71(3):495-548. doi:10.1128/MMBR.00005-07.
- Collins SM, Bercik P. The Relationship Between Intestinal Microbiota and the Central Nervous System in Normal Gastrointestinal Function and Disease. Gastroenterology. 2009;136(6):2003-2014. doi:10.1053/j.gastro.2009.01.075.
- Live S, Microbiome T, State N. Microbiome and Neuroscience : The Mind-bending Power of Bacteria | The Kavli Foundation. 2015:1-7.
- Erejuwa OO, Sulaiman S a., Ab Wahab MS. Modulation of gut microbiota in the management of metabolic disorders: The prospects and challenges. Int J Mol Sci. 2014;15(3):4158-4188. doi:10.3390/ijms15034158.
- Collins SM, Surette M, Bercik P. The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol. 2012;10(11):735-742. doi:10.1038/nrmicro2876.
- Schnorr SL, Candela M, Rampelli S, et al. Gut microbiome of the Hadza hunter-gatherers. Nat Commun. 2014;5:3654. doi:10.1038/ncomms4654.
- Lyte M. Microbial Endocrinology in the Microbiome-Gut-Brain Axis: How Bacterial Production and Utilization of Neurochemicals Influence Behavior. PLoS Pathog. 2013;9(11):e1003726. doi:10.1371/journal.ppat.1003726.
- Geuking MB, Köller Y, Rupp S, McCoy KD. The interplay between the gut microbiota and the immune system. Gut Microbes. 2014;5(3). doi:10.4161/gmic.29330.
- Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13(10):701-712. doi:10.1038/nrn3346.
- Villiger PM. Interactions between the nervous and the immune system. Schweiz Med Wochenschr. 1994;124(20):857-866. doi:10.1016/S0140-6736(95)90066-7.
- Shoaie S, Karlsson F, Mardinoglu A, Nookaew I, Bordel S, Nielsen J. Understanding the interactions between bacteria in the human gut through metabolic modeling. Sci Rep. 2013;3:2532. doi:10.1038/srep02532.
- Greve MW, Zink BJ. Pathophysiology of Traumatic Brain Injury Address Correspondence to. Medicine (Baltimore). 2009;76(2):97-104. doi:10.1002/MSJ.
- 16S rRNA from Human Neonatal Microbiome in Necroti… (ID 46337) – BioProject – NCBI. http://www.ncbi.nlm.nih.gov/bioproject/46337. Accessed April 23, 2015.
- 16S rRNA from Human Gut Microbiome in Children wit… (ID 46339) – BioProject – NCBI. http://www.ncbi.nlm.nih.gov/bioproject/46339. Accessed April 23, 2015.
- Human Vaginal Microbiome (ID 46877) – BioProject – NCBI. http://www.ncbi.nlm.nih.gov/bioproject/46877. Accessed April 23, 2015.
- Abou-Donia MB, El-Masry EM, Abdel-Rahman A a, McLendon RE, Schiffman SS. Splenda alters gut microflora and increases intestinal p-glycoprotein and cytochrome p-450 in male rats. J Toxicol Environ Health A. 2008;71(21):1415-1429. doi:10.1080/15287390802328630.
- Kim YH, Kwon HS, Kim DH, et al. Piceatannol, a stilbene present in grapes, attenuates dextran sulfate sodium-induced colitis. Int Immunopharmacol. 2008;8(12):1695-1702. doi:10.1016/j.intimp.2008.08.003.
- 16S rRNA from Human Gut Microbiome in Ulcerative C… (ID 46315) – BioProject – NCBI. http://www.ncbi.nlm.nih.gov/bioproject/46315. Accessed April 23, 2015.
- DiBaise JK, Frank DN, Mathur R. Impact of the Gut Microbiota on the Development of Obesity: Current Concepts. Am J Gastroenterol Suppl. 2012;1(1):22-27. doi:10.1038/ajgsup.2012.5.
- Fasano A. Zonulin and its regulation of intestinal barrier function: the biological door to inflammation, autoimmunity, and cancer. Physiol Rev. 2011;91(1):151-175. doi:10.1152/physrev.00003.2008.
- Human Virome in Febrile Illness (ID 46335) – BioProject – NCBI. http://www.ncbi.nlm.nih.gov/bioproject/46335. Accessed April 23, 2015.
- Dethlefsen L, McFall-Ngai M, Relman D a. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449(7164):811-818. doi:10.1038/nature06245.
- Collado MC, Rautava S, Isolauri E, Salminen S. Gut microbiota: a source of novel tools to reduce the risk of human disease? Pediatr Res. 2014;77(1-2):182-188. doi:10.1038/pr.2014.173.
- Lee HC, Jenner AM, Low CS, Lee YK. Effect of tea phenolics and their aromatic fecal bacterial metabolites on intestinal microbiota. Res Microbiol. 2006;157(9):876-884. doi:10.1016/j.resmic.2006.07.004.
- Gut A, Gut A, Gut A, Gut A, Gut A, Project HM. Preliminary Characterization of the American Gut Population Who is participating in the AGP ? Our participants Vitamin use. 2014;3. https://www.microbio.me/AmericanGut/static/img/mod1_main.pdf.
- Human Gut Microbiome in Crohn’s Disease (ID 46879) – BioProject – NCBI. http://www.ncbi.nlm.nih.gov/bioproject/46879. Accessed April 23, 2015.
- Peterson G, Kumar A, Gart E, Narayanan S. Catecholamines increase conjugative gene transfer between enteric bacteria. Microb Pathog. 51(1-2):1-8. doi:10.1016/j.micpath.2011.03.002.
- Magrone T, Jirillo E. The interplay between the gut immune system and microbiota in health and disease: nutraceutical intervention for restoring intestinal homeostasis. Curr Pharm Des. 2013;19(7):1329-1342. http://www.ncbi.nlm.nih.gov/pubmed/23151182.
- Nelson KE, Weinstock GM, Highlander SK, et al. A catalog of reference genomes from the human microbiome. Science. 2010;328(5981):994-999. doi:10.1126/science.1183605.
- Dupaul-Chicoine J, Dagenais M, Saleh M. Crosstalk between the intestinal microbiota and the innate immune system in intestinal homeostasis and inflammatory bowel disease. Inflamm Bowel Dis. 2013;19(10):2227-2237. doi:10.1097/MIB.0b013e31828dcac7.
- Fall B, Litchy A, Schafer M. Microbiome. 2014.
- Dinan TG, Stilling RM, Stanton C, Cryan JF. Collective unconscious: how gut microbes shape human behavior. J Psychiatr Res. 2015:1-9. doi:10.1016/j.jpsychires.2015.02.021.
- 16S rRNA from Human Vaginal Microbiome (ID 46311) – BioProject – NCBI. http://www.ncbi.nlm.nih.gov/bioproject/46311. Accessed April 23, 2015.
- 16S rRNA from Human Gut Microbiome in Amish Obesity (ID 46319) – BioProject – NCBI. http://www.ncbi.nlm.nih.gov/bioproject/46319. Accessed April 23, 2015.
- National Institutes of Health. Human Microbiome Project. 2013. http://commonfund.nih.gov/hmp/. Accessed April 22, 2015.
- 16S rRNA from Human Gut Microbiome in Crohn’s Dise… (ID 46313) – BioProject – NCBI. http://www.ncbi.nlm.nih.gov/bioproject/46313. Accessed April 23, 2015.
- Heijtz RD, Wang S, Anuar F, et al. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci U S A. 2011;108(7):3047-3052. doi:10.1073/pnas.1010529108.
- Sutherland B a., Rahman RM a, Appleton I. Mechanisms of action of green tea catechins, with a focus on ischemia-induced neurodegeneration. J Nutr Biochem. 2006;17(5):291-306. doi:10.1016/j.jnutbio.2005.10.005.
- Functional Gene Metagenome from Human Gut Microbio… (ID 50637) – BioProject – NCBI. http://www.ncbi.nlm.nih.gov/bioproject/50637. Accessed April 23, 2015.
- Ridaura VK, Faith JJ, Rey FE, et al. Gut Microbiota from Twins Discordant for Obesity Modulate Metabolism in Mice Gut Microbiota from Twins Metabolism in Mice. Science. 2013;341(September):1241214. doi:10.1126/science.1241214.
- 16S rRNA from Human Cutaneous Microbiome in Psoria… (ID 46309) – BioProject – NCBI. http://www.ncbi.nlm.nih.gov/bioproject/46309. Accessed April 23, 2015.
- Lane DJ, Pace B, Olsen GJ, Stahl D a, Sogin ML, Pace NR. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A. 1985;82(20):6955-6959. doi:10.1073/pnas.82.20.6955.
- 16S rRNA from Human Foregut Microbiome in Esophage… (ID 46307) – BioProject – NCBI. http://www.ncbi.nlm.nih.gov/bioproject/46307. Accessed April 23, 2015.
- Human Gut Microbiome in Ulcerative Colitis (ID 46881) – BioProject – NCBI. http://www.ncbi.nlm.nih.gov/bioproject/46881. Accessed April 23, 2015.
- 16S rRNA from Human Urethral Microbiome in Adolesc… (ID 46317) – BioProject – NCBI. http://www.ncbi.nlm.nih.gov/bioproject/46317. Accessed April 23, 2015.
- Zipris D. The interplay between the gut microbiota and the immune system in the mechanism of type 1 diabetes. Curr Opin Endocrinol Diabetes Obes. 2013;20(4):265-270. doi:10.1097/MED.0b013e3283628569.
- 16S rRNA from Human Vaginal Microbiome in Bacteria… (ID 46331) – BioProject – NCBI. http://www.ncbi.nlm.nih.gov/bioproject/46331. Accessed April 23, 2015.
- Sergent T, Piront N, Meurice J, Toussaint O, Schneider YJ. Anti-inflammatory effects of dietary phenolic compounds in an in vitro model of inflamed human intestinal epithelium. Chem Biol Interact. 2010;188(3):659-667. doi:10.1016/j.cbi.2010.08.007.
- Ames BN, Wakimoto P. Are vitamin and mineral deficiencies a major cancer risk? Nat Rev Cancer. 2002;2(9):694-704. doi:10.1038/nrc886.
- Marques TM, Cryan JF, Shanahan F, et al. Gut microbiota modulation and implications for host health: Dietary strategies to influence the gut-brain axis. Innov Food Sci Emerg Technol. 2014;22:239-247. doi:10.1016/j.ifset.2013.10.016.
- Stokes BE, Response I. Micro Flora : A Community of Wellness. 2015:20-22.
- Lyte M, Ernst S. Catecholamine induced growth of gram negative bacteria. Life Sci. 1992;50(3):203-212. http://www.ncbi.nlm.nih.gov/pubmed/1731173. Accessed April 23, 2015.
- Human Gut Microbiome in Crohn’s Disease (ID 46321) – BioProject – NCBI. http://www.ncbi.nlm.nih.gov/bioproject/46321. Accessed April 22, 2015.
- IMG Data Management & Analysis Systems. Http://ImgJgiDoeGov. http://img.jgi.doe.gov/. Accessed April 22, 2015.
- Marín M, María Giner R, Ríos JL, Carmen Recio M. Intestinal anti-inflammatory activity of ellagic acid in the acute and chronic dextrane sulfate sodium models of mice colitis. J Ethnopharmacol. 2013;150(3):925-934. doi:10.1016/j.jep.2013.09.030.
- 16S rRNA from Human Skin Microbiome Associated wit… (ID 46327) – BioProject – NCBI. http://www.ncbi.nlm.nih.gov/bioproject/46327. Accessed April 23, 2015.
- Li J, Jia H, Cai X, et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32(8):834-841. doi:10.1038/nbt.2942.
- Rosillo MA, Sánchez-Hidalgo M, Cárdeno A, et al. Dietary supplementation of an ellagic acid-enriched pomegranate extract attenuates chronic colonic inflammation in rats. Pharmacol Res. 2012;66(3):235-242. doi:10.1016/j.phrs.2012.05.006.
- Sánchez-Fidalgo S, Cárdeno a., Sánchez-Hidalgo M, et al. Dietary unsaponifiable fraction from extra virgin olive oil supplementation attenuates acute ulcerative colitis in mice. Eur J Pharm Sci. 2013;48(3):572-581. doi:10.1016/j.ejps.2012.12.004.
- O’Mahony SM, Felice VD, Nally K, et al. Disturbance of the Gut microbiota in early-life selectively affects visceral pain in adulthood without impacting cognitive or anxiety-related behaviors in male rats. Neuroscience. 2014;277:885-901. doi:10.1016/j.neuroscience.2014.07.054.
- Hur SJ, Kang SH, Jung HS, et al. Review of natural products actions on cytokines in inflammatory bowel disease. Nutr Res. 2012;32(11):801-816. doi:10.1016/j.nutres.2012.09.013.
- Cryan JF, O’Mahony SM. The microbiome-gut-brain axis: From bowel to behavior. Neurogastroenterol Motil. 2011;23(3):187-192. doi:10.1111/j.1365-2982.2010.01664.x.
- human gut metagenome (ID 174514) – BioProject – NCBI. http://www.ncbi.nlm.nih.gov/bioproject/174514. Accessed April 23, 2015.
- Grundmann O, Yoon SL. Complementary and alternative medicines in irritable bowel syndrome: an integrative view. World J Gastroenterol. 2014;20(2):346-362. doi:10.3748/wjg.v20.i2.346.
- Mochizuki M, Hasegawa N. Therapeutic efficacy of pycnogenol in experimental inflammatory bowel diseases. Phyther Res. 2004;18(12):1027-1028. doi:10.1002/ptr.1611.
- O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7(7):688-693. doi:10.1038/sj.embor.7400731.
- Dossier P. ENZO : PROFESSIONAL Product Dossier. 2010;(September).
- 16S rRNA from Human Gut Microbiome in Crohn’s Dise… (ID 46323) – BioProject – NCBI. http://www.ncbi.nlm.nih.gov/bioproject/46323. Accessed April 23, 2015.
- Human Skin Microbiome in Disease States (ID 46333) – BioProject – NCBI. http://www.ncbi.nlm.nih.gov/bioproject/46333. Accessed April 23, 2015.
- Health M. Probiotics for the Mind A Crossroads of the Microbiome and Mental Health Table of Contents.
- Goodrich KM, Fundaro G, Griffin LE, et al. Chronic administration of dietary grape seed extract increases colonic expression of gut tight junction protein occludin and reduces fecal calprotectin: A secondary analysis of healthy Wistar Furth rats. Nutr Res. 2012;32(10):787-794. doi:10.1016/j.nutres.2012.09.004.
- Suez J, Korem T, Zeevi D, et al. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature. 2014. doi:10.1038/nature13793.
- Ley RE. Obesity and the human microbiome. Curr Opin Gastroenterol. 2010;26(1):5-11. doi:10.1097/MOG.0b013e328333d751.